Glycomics is the comprehensive study of all glycans expressed in biological systems. Glycan markers hold considerable opportunities and challenges for disease diagnosis because the glycosylation machinery is highly sensitive to the biochemical environment. The development of new analytical methods has significantly increased the progress in understanding the role of the glycome in many biological areas. Research in our group has focus on glycans as disease markers. The detection and analysis of the glycans that decorate the underlying proteins may provide more specific detection of cancer rather than examining the protein themselves. Proteins are often modified by the attachment of glycans during the normal course of protein production. It is estimated that over 50% of all human proteins are modified in this way. The biosynthesis of glycan relies on a number of highly competitive processes involving glycosyl transferase, suggesting that the expression of the glycan products is highly variable. There are many studies demonstrating that the glycans produced in the cancer cells are significantly different from those in normal cells. Aberrant glycosylation is also observed with many other diseases.
In the studies in our laboratory, we harvest glycans from human fluids such as serum, tear, and saliva to determine whether the glycosylation has changed in cancer patients compared to healthy controls. This new glycan assay is used to discover biomarkers in the diagnosis of cancer. Global glycan profiling of human serum based on mass spectrometry has already led to several potentially promising markers for several types of cancer and diseases. In addition a method to assess the diversity (specific isomer structures) of the N-glycans in human serum without derivatization has been developed by our laboratory employing porous graphitized carbon as stationary phase. The ability to separate and simultaneously analyze neutral and anionic N-glycans from human serum without derivatization demonstrates a rapid yet highly sensitive and highly specific tool for disease biomarker discovery.

Cancer biomarkers
Glycosylation is an important determinant of protein function and has been shown to mediate many cancer-related processes and has recently been acknowledged as potential markers for diseases.
Show below are average relative abundances and standard error bars associated with five structural isomers of Hex5HexNAc4FucNeuAc in ovarian cancer cases (striped pink, n=48) and controls (checkered blue, n = 46). *Statistically significant difference between ovarian cancer cases and controls.
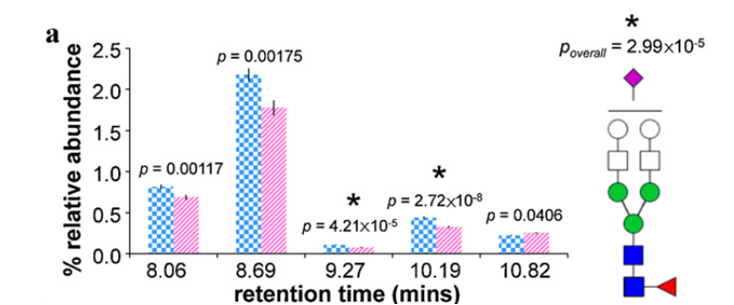
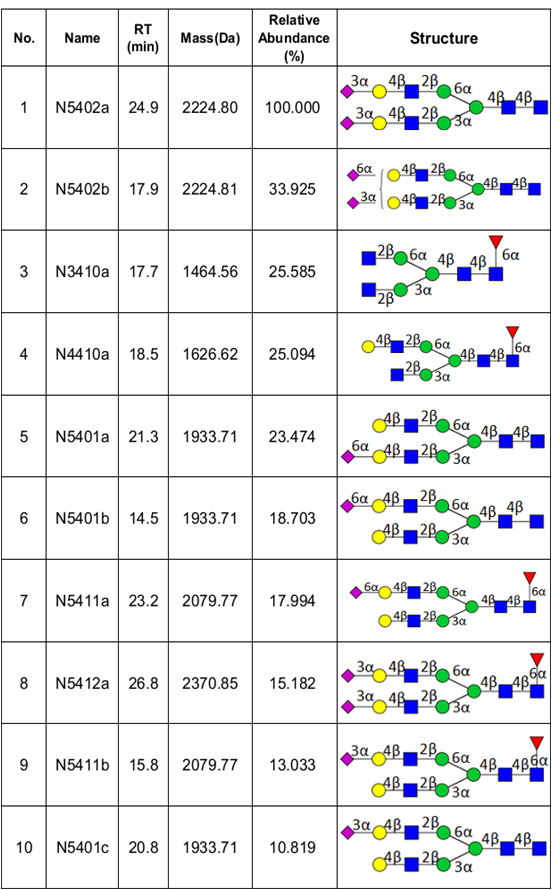
We have recently optimized a rapid-throughput method for comprehensive, isomer-specific chromatographic profiling of native serum glycans and applied it to glycan biomarker discovery. In the near future we will apply a rapidly developing structural serum N-glycan library featuring isomers with identifiers, relative abundances, retention times, accurate masses, and tandem MS spectra (below).
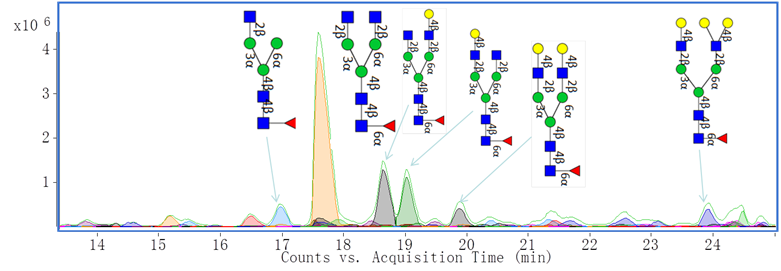
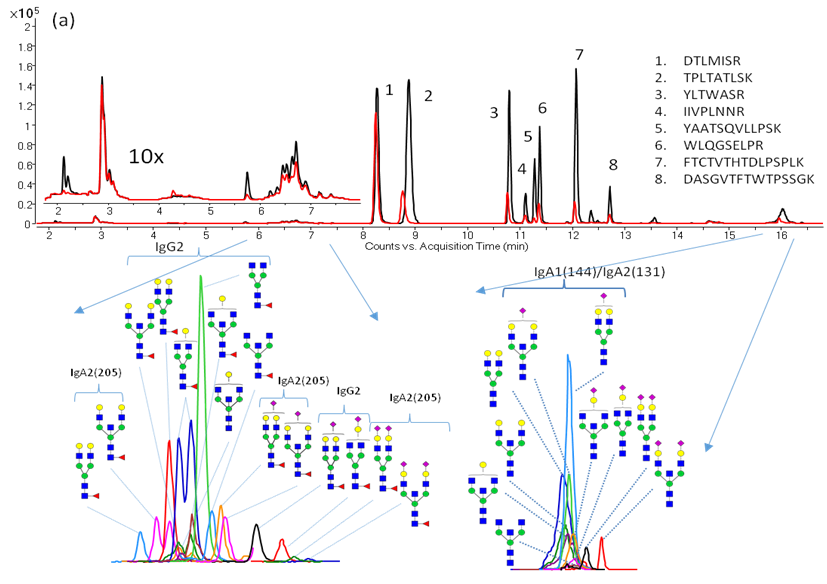
Additionally, we have characterized monoclonal antibody (mAb) glycan profiles. Glycosylation may affect the therapeutic efficacy, pharmacokinetics, immunogenicity, folding and stability of these proteins. Shown below is an annotated extracted compound chromatogram of Eculizumab.
Cell Surface Glycosylation
Glycosylation on the cell surface membrane plays an important role in cell signaling and bioactivity. We have optimized a method to isolate and enrich for cell membrane glycans and use analytical tools to study them. In recent studies, we profiled cell membrane N-glycans from a variety of cell lines, and found that the same types displayed similar trends. Graphed below is a color-coded representation of the Pearson correlation coefficient (R) between each pair of cell lines (Red- high correlation, blue- low correlation). Relative abundances are shown for each glycan glass. ES-2 is clearly distinct from the group of lymphoma cell lines.
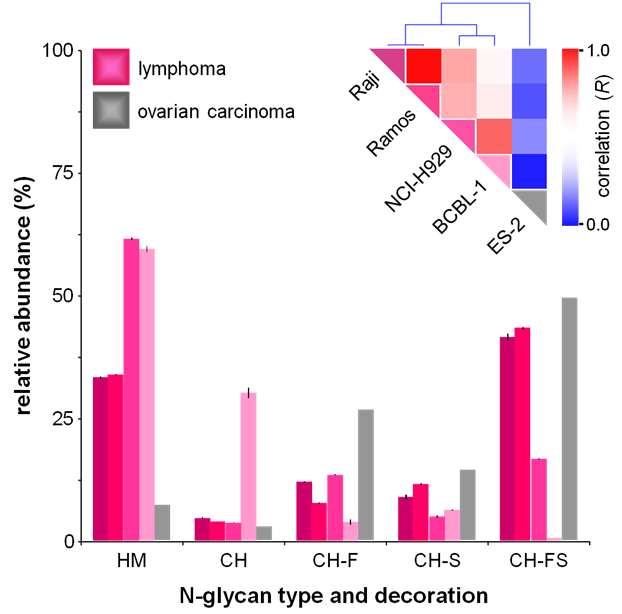
Above: Glycan class profiles of four B-cell lymphoma cell lines: Raji, Ramos, NCI-H929, BCBL-1 and one ovarian carcinoma cell line (ES-2) analyzed by HPLC Chip/TOF-MS.
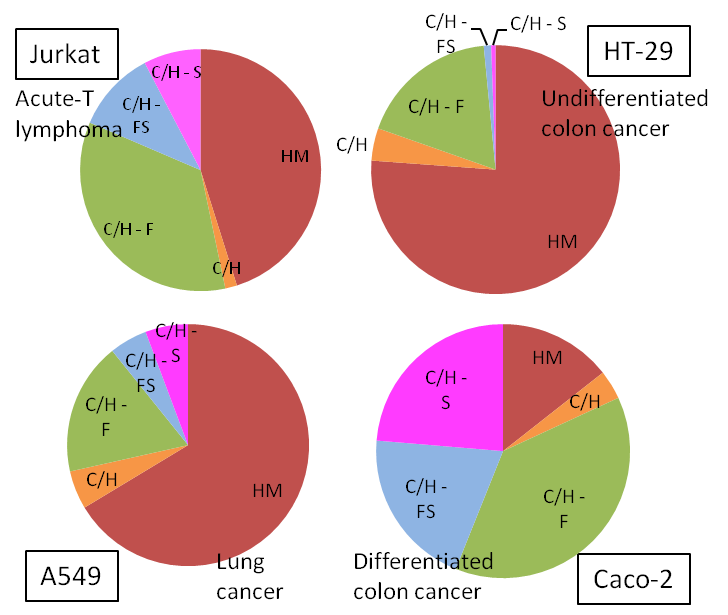
Four different cancerous cell lines (shown below) were analyzed by HPLC Chip/TOF-MS. Each cancer type depicted a unique N-glycan profile. The pie charts below display average triplicate relative abundances of N-glycans from each cell line. HM- High Mannose, C/H- Complex/Hybrid, CH-F- Complex/Hybrid Fucosylated, C/H-FS- Complex/Hybrid Fucosylated Sialylated, C/H-S- Complex/Hybrid Sialylated
Markers for Autoimmunity
Over the last decades, several successful studies have emerged on the detection of candidate glycan biomarkers for several diseases including auto-immune diseases. Most studies have focused on whole biofluids, and changes in specific glycoforms of proteins may result in more important biomarkers, with improved sensitivity and specificity. Here, we present methods to investigate protein- and site- specific glycan profiles of the immunoglobulins A, G, and M in a quantitative manner.
Below: Simultaneous absolute immunoglobulin quantitation and glycosylation analysis directly from serum allows us to apply this method to clinical studies.
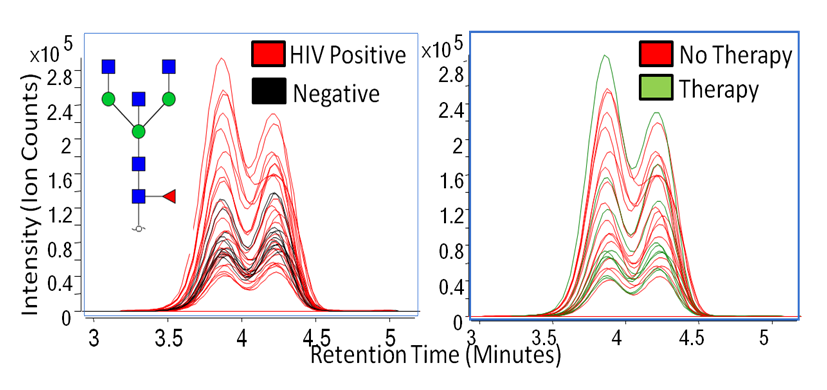
Human immunodeficiency virus (HIV) has been known to weaken the immune system, and many studies have proven that immunoglobulin activity is known to fluctuate from patient to patient. Because glycomic profiling of human serum and site-specific protein quantitation in HIV has not been thoroughly examined, we have compared the glycosylation profiles between four types of HIV patient groups to determine whether or not immunoglobulin G (IgG) glycosylation varies in these patients. Shown below are overlaid extracted compound chromatograms of Hex3HexNAc5Fuc1 found in HIV positive and controls (left) and therapy and no therapy groups (right).
Below: Differences in the glycosylation patterns of all immunoglobulins can be observed between the patient sets (Negative, Therapy, Long-term no therapy, No Therapy). The glycopeptide response was normalized to remove the contribution of protein concentrations. This allowed for comparison of only the glycosylation profiles.
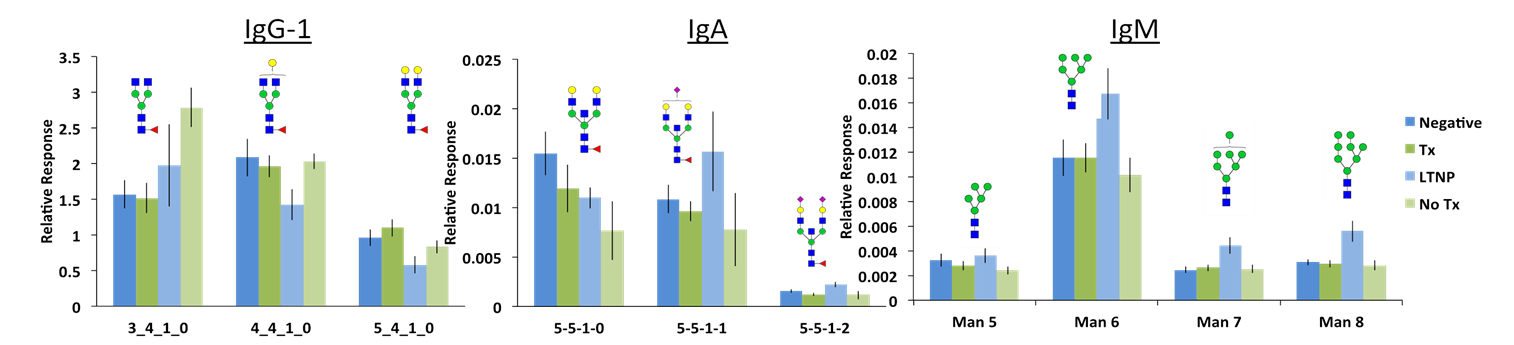
References:
- Lebrilla CB, An HJ (2009) The prospects of glycan biomarkers for the diagnosis of diseases. Molecular bioSystems 5 (1):17-20. doi:10.1039/b811781k
- Hua S, Saunders M, Dimapasoc LM, Jeong SH, Kim BJ, Kim S, So M, Lee KS, Kim JH, Lam KS, Lebrilla CB, An HJ (2013) Differentiation of Cancer Cell Origin and Molecular Subtype by Plasma Membrane N-Glycan Profiling. Journal of proteome research. doi:10.1021/pr400987f
- Song T, Aldredge D, Lebrilla CB (2015) A method for in-depth structural annotation of human serum glycans yields the biological variations. Analytical chemistry 87 (15):7754-7762. doi:10.1021/acs.analchem.5b01340
- Song T, Ozcan S, Becker A, Lebrilla CB (2014) In-Depth Method for the Characterization of Glycosylation in Manufactured Recombinant Monoclonal Antibody Drugs.. Analytical chemistry 86 (12):5661-5666. doi:10.1021/ac501102t
- Park D, Brune KA, Mitra A, Marusina AI, Maverakis E, Lebrilla CB (2015) Characteristic changes in cell surface glycosylation accompany intestinal epithelial cell differentiation: high mannose structures dominate the cell surface glycome of undifferentiated enterocytes. Molecular & cellular proteomics : MCP. doi:10.1074/mcp.M115.053983
- Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, Lebrilla CB (2011) Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers.. The Analyst 136 (18):3663-3671. doi:10.1039/c1an15093f
- Ruhaak LR, Barkauskas DA, Torres J, Cooke CL, Wu LD, Stroble C, Ozcan S, Williams CC, Camorlinga M, Rocke DM, Lebrilla CB, Solnick JV (2015) The serum immunoglobulin G glycosylation signature of gastric cancer. EuPA Open Proteom 6:1-9. doi:10.1016/j.euprot.2014.11.002
- Ruhaak LR, Lebrilla CB (2015) Applications of Multiple Reaction Monitoring to Clinical Glycomics 78 (5-6):335-342. doi:10.1007/s10337-014-2783-9
- Ruhaak LR, Taylor SL, Stroble C, Nguyen UT, Parker EA, Song T, Lebrilla CB, Rom WN, Pass H, Kim K, Kelly K, Miyamoto S (2015) Differential N-glycosylation patterns in lung adenocarcinoma tissue.. doi:10.1021/acs.jproteome.5b00255